Overview
The UCLA Center for Tropical Research (CTR) is at the forefront of research and surveillance of avian influenza virus (bird flu or avian flu) in wild birds. CTR has led avian influenza virus (AIV) surveys of migratory land birds in both North America and Central Africa. Recently, CTR was awarded a four-year project from the National Institutes of Health/National Institute of Allergy and Infectious Diseases (NIH/NIAID) entitled “Effects of Avian Migration and Anthropogenic Change on the Distribution and Transmission Risks of Avian Influenza.”
With this generous support, we are conducting research to determine the role that North American migratory birds play in the dispersion of AIV strains between breeding sites in Canada and the U.S. and wintering sites in Mexico, Central America, and South America. In addition, we plan to investigate the impact that human land use changes may have on migratory routes and the potential transmission of influenza between hosts, particularly between humans and birds.
Research
Our research is divided into four parts:
- Identifying the distribution and prevalence of avian influenza strains in Nearctic-Neotropical migratory birds.
- Establishing the extent to which AIV distributions correlate with known patterns of migratory connectivity.
- Determining the incidence of AIV transmission from birds to humans and whether the strain distributions in humans reflect the strain distributions in birds.
- Identifying, using remote sensing data, the environmental variables that are associated with AIV strain distribution. This data will be incorporated into ecological models used to predict how human changes in habitat can affect transmission dynamics between migratory species, between migratory and resident species, and between birds and humans.
These studies take advantage of our unique collaboration with the major bird banding station networks in the Americas, our extensive experience in avian field research in Central Africa, and UCLA’s unique resources and expertise in infectious diseases.
1a. Summary of Avian Influenza
Avian influenza is a disease caused by the influenza A virus, an RNA virus of the family Orthomyxoviridae (‘myxo’ is derived from the Greek word for mucous). Three types of influenza viruses (A, B, and C) are known. These viruses are classified based on the antigenic properties of two viral proteins (matrix and nucleoprotein). Influenza A is capable of infecting birds and mammals (including humans), whereas Influenza B and C viruses are only known to infect mammals (including humans).
Avian influenza virus (AIV) is a natural disease in wild birds, and all known subtypes of influenza A have been isolated from wild birds. AIV is generally carried in the gastrointestinal tract and is spread via feces, saliva, and nasal secretions. Most species, however, do not exhibit symptoms of infection. Poultry and captive birds are more susceptible to AIV and tend to manifest disease symptoms.
There are two basic forms of AIV in bird populations: low-pathogenic (LPAIV) and high-pathogenic (HPAIV). LPAIV is the most common form of virus found in wild birds. Domestic birds, when infected with LPAIV, exhibit mild symptoms of disease. HPAIV, on the other hand, is not common in wild birds. Domesticated flocks, however, are particularly susceptible to these strains and experience high mortality. HPAIV is responsible for considerable economic losses, international trade sanctions, and human mortality. Examples of HPAIV include the variants of H5N1 currently circulating in Asia, Africa, and Europe.
1b. Influenza A and Humans

Seasonal flu in humans is caused by a subset of influenza A subtypes (H3N2, H2N2, H1N1, and H1N2). These viruses can cause respiratory illness with symptoms ranging from mild to severe. These infections can be passed from human to human via breathing airborne virus particles.
Avian flu in humans is much less common. If a human is infected with AIV the symptoms can range from those typical to seasonal flu to a life-threatening illness. The extent to which humans contract LPAIV is unknown, but since 1997 approximately 200 cases of HPAIV H5N1 have been reported.
Transmission of AIV from birds to humans requires a fecal oral route, meaning that a human needs to ingest virus shed from a bird in order to be infected. This can occur if the human lives or works in close proximity to birds or comes in contact with a contaminated surface. Fortunately, human-to-human transmission of AIV has not been reported. However, AIV is a rapidly evolving group virus; if a strain of HPAIV were to gain the ability to transmit between humans, then it is probable that pandemic flu will occur.
Pandemic flu, or a worldwide epidemic of flu, is characterized as a severe disease caused by a novel strain of influenza that is transmissible between people. Populations of humans are highly susceptible to infection by novel strains of flu because they have little or no immunity to these strains. During the 20th century, three separate influenza pandemics occurred. The most devastating of these was the Spanish flu pandemic of 1918, which took the lives of over 50 million people.
1c. Influenza A Biology
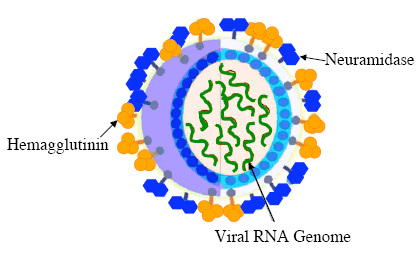
The influenza A virion (virus particle) is generally spherical and between 80-120 nm in diameter. Its genome is made up of eight different pieces of single-stranded RNA, which encode 11 different viral proteins. Of these proteins, the best characterized are the hemagglutinin (H or HA) and neuraminidase (N or NA) glycoproteins. Both of these proteins are found on the outside of the virion and are important in infecting host cells and host antigenic response. Influenza viral subtypes (or serotypes) are identified by the immunological response of antibodies to the combination of H and N proteins on the viral coat.
Influenza A is a rapidly evolving virus because the genome is continually accumulating point mutations. These mutations are due to the enzyme that is used to replicate the viral genome. This enzyme lacks a proofreading mechanism. This continuous change allows strains of the virus to routinely infect humans even if they have been infected by a similar strain in the past. In terms of evading immune response, the most important virus mutations occur on the H and N surface proteins. The two distinct ways in which influenza A changes its genetic composition and effectively outsmarts the immune system are through antigenic drift and antigenic shift.
Antigenic drift refers to the gradual accumulation of point mutations in the genes that code for the H and N surface proteins. Over time, mutations will accumulate to the point that the specific immunity that a host had developed to an ancestral strain will be insufficient to defend against the evolved descendent of that virus.
Antigenic shift, on the other hand, is when two or more viruses infect the same cell and swap genetic material. This swapping creates a virus with a novel subtype; this involves major rearrangements of the original H and N surface proteins. Human populations typically have little to no immune protection against the novel strains generated from antigenic shift, making them highly susceptible to infection.
For more information, please visit the following web pages:
- www.influenzareport.com
- www.cdc.gov/flu/avian
- www.who.int/csr/disease/avian_influenza/en
- http://en.wikipedia.org/wiki/Avian_influenza
- http://en.wikipedia.org/wiki/Spanish_flu
2a. Summary of Migratory Connectivity
Migratory connectivity is a term used to describe the relationship between populations of animals (especially birds) and geographic locations at different time points during the year. North American migratory birds typically breed in northern latitudes during the summer and overwinter in southern latitudes. In general, populations of a migratory species will take one of four major migratory routes between breeding and wintering grounds. However, not all populations of a species take the same routes. For example, Swainson’s thrush (Catharus ustulatus) take two different non-overlapping migratory routes to and from northern and southern latitudes. Coastal populations travel along the Pacific coast while continental populations migrate along eastern routes (Reugg and Smith 2002).
Migratory connectivity studies are particularly useful for developing conservation strategies, tracking bird-borne diseases, and for understanding population genetics, gene flow, and speciation. Advances in genetic technologies and stable-isotope analysis are increasing our ability to detect patterns of migratory connectivity. Traditional methods of banding and recapture provide limited details about the migratory patterns of a species of bird because few banded individuals are ever captured twice. However, genetic techniques can provide population-level data about breeding and wintering grounds and the migratory routes connecting the two.
Genetic analyses take advantage of population-specific variations in the DNA of bird species. Members of the same population are more closely related and, therefore, have more genetic markers in common than individuals from two different populations. If a species of bird maintains several different breeding populations, then the genetic differences between populations can be used to connect individual birds back to a specific population. Thus, capturing individual birds at any point during the year can provide information about where a specific population breeds, and overwinters, and which migratory routes it follows.
Stable-isotopic analysis can also be used to help identify the breeding grounds used by individual birds. There are known longitudinal and latitudinal concentration gradients for stable isotopes. Animals feeding at a location will ingest food with a specific concentration of stable isotope. These isotopes will then be incorporated into the physical structures of the bird that are developed at that location (e.g., feathers). Thus one can estimate the location where feathers are developed by the concentrations of stable isotope present in those feathers. If a bird is captured on its wintering grounds, the regional location of the breeding grounds from the previous summer can be identified.
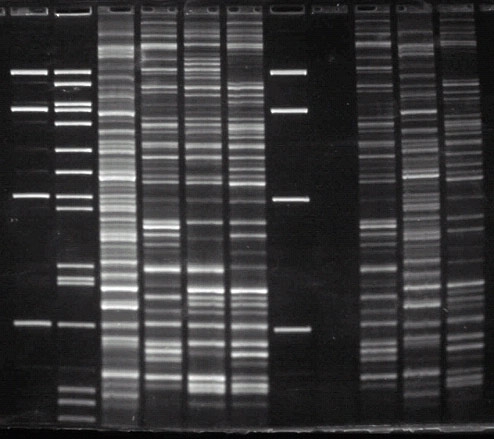
This image of an AFLP (Amplified Fragment Length Polymorphism) assay is an example of a genetic fingerprinting method that is used to assess migratory connectivity.
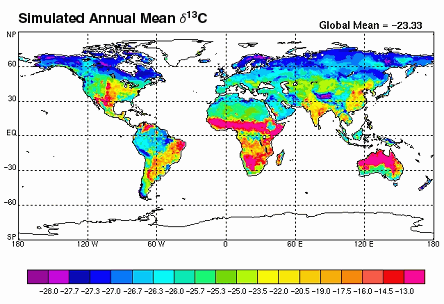
This image (from Biocycle) is an example of a stable isotope concentration gradient map.
2b. Avian Influenza A and Migratory Birds
Bird flu reached the public’s attention in 1996 and 1997 after the H5N1 outbreaks of highly virulent avian influenza A virus (AIV) in Asia, Africa, and Europe. Researchers have known for years that low pathogenic AIV is prevalent in birds. In fact, AIV has been isolated from dozens of species. Despite this knowledge, little is actually known about the prevalence of AIV in birds or how AIV is transmitted through avian populations via migratory routes (Stallknecht and Shane 1988; Webster et al.1992). The threat of H5N1 in migratory species has made this field of study particularly relevant.
Several lineages of H5N1 have been found in poultry, and data suggests that H5N1 evolved in several hosts. Historically, the first H5 avian subtype was identified from wild terns in South Africa in 1961 (A/tern/South Africa/61 [H5N3]), and the precursor of the current highly pathogenic H5N1 lineage was sampled in China around 1996. Since then, numerous genotypes of H5N1 have circulated throughout Southeast Asia via waterfowl and poultry (Webster et al. 2006 ).
The H5N1 viruses probably emerged in Asia in 2001 and 2002 from the reassortment and evolution of many different genotypes. These putative ancestor genotypes were detected in poultry markets and farms in Hong Kong (Li et al. 2004; Chen et al. 2005; Liu et al. 2005). In August 2005, highly pathogenic H5N1 killed thousands of migratory waterfowl around Lake Qinghai in western China (Liu et al. 2005 ) and an additional 89 migratory birds in Mongolia. By February of 2006, H5N1 was isolated from 21 wild bird species (mostly in Asia) and 23 captive species. An additional 12 species have been experimentally infected.
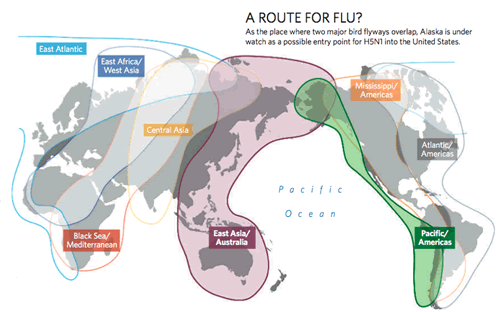
Although major outbreaks of H5N1 occurred independent of typical migration cycles, migratory birds have been strongly implicated in the carriage and dissemination of the H5N1 strain. To date several passerine species have been identified as potential conduits for the introduction of AIV subtypes, including H5N1, from Asia to North America. For example, Yellow Wagtails commonly breed in western Alaska and winter near agricultural fields in southeast China and Indonesia (Badyaev et al. 1998). Some North American Nearctic-Neotropical migrant land birds share breeding and/or migration stopover habitats with Yellow Wagtails and other Eurasian migratory species that could potentially carry a variant of H5N1.
The information regarding the dissemination of AIV along migratory routes, and between birds and humans, will be critical if H5N1 enters populations of North American migratory birds. Some species of non-migratory birds (e.g., Sparrows, Starlings, Blackbirds, and nest-parasitizing Cowbirds) share the same habitat as poultry and domesticated birds. If these non-migratory birds contract H5N1 from carrier migratory birds, then they could spread the strain to poultry. Poultry are very susceptible to H5N1 and could, in turn, infect humans associated with the poultry trade. Thus, non-migratory birds and poultry would act as vectors for the transmission of AIV between migratory birds and humans.
A compounding factor of AIV detection in wild birds is that it tends to asymptomatically infect songbirds. This means that an H5N1 virus could potentially spread and evolve in bird populations and along migratory routes with little warning or initial indication. Widespread monitoring of North American land birds is paramount. Not only is it important for the detection of H5N1, but also for providing details about the ecology of endemic low-pathogenic AIV. Ecological information could then be used to generate a predictive model of a potential H5N1 outbreak.
2c. References
Badyaev, A. V., B. Kessel, and D. D. Gibson. 1998. Yellow Wagtail (Motacilla flava). In A. Poole and F. Gill (Eds.) The Birds of North America, No. 382. The Birds of North America, Inc., Philadelphia, PA.
Chen, H., G. J. D. Smith, S. Y. Zhang, K. Qin, J. Wang, K. S. Li, R. G. Webster, J. S. M. Peiris, and Y. Guan. 2005. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature 436: 191–2.
Li, K. S., Y. Guan, J. Wang, G. J. Smith, K. M. Xu, L. Duan, A. P. Rahardjo, P. Puthavathana, C. Buranathai, T. D. Nguyen, A. T. S. Estoepangestie, A. Chaisingh, P. Auewarakul, H. T. Long, N. T. H. Hanh, R. J. Webby, L. L. M. Poon, H. Chen, K. F. Shortridge, K. Y. Yuen, R. G. Webster, and J. S. M. Peiris. 2004. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430: 209–213.
Liu, J., H. Xiao, F. Lei, Q. Zhu, K. Qin, X. W. Zhang, X. L. Zhang, D. Zhao, G. Wang, Y. Feng, J. Ma, W. Lui, J. Wang, and G. F. Gao. 2005. Highly pathogenic H5N1 influenza virus infection in migratory birds. Science 309: 1206.
Ruegg, K. and T. B. Smith. 2002. Not as the crow flies: a historical explanation for circuitous migration in Swainson’s thrush (Catharus ustulatus). Proceedings of the Royal Society, London 269: 1375-1381.
Stallknecht, D. E. S. and M. Shane. 1998. Host range of avian influenza virus in free-living birds. Veterinary Research Communications 12: 125–141.
Webster, R. G , W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiology Review 56: 152-179.
Webster, R. G., M. Peiris, H. Chen, and Y. Guan. 2006. H5N1 Outbreaks and Enzootic Influenza. Emerging Infectious Diseases 12: 3-8.
Collaborating Organizations
Collaborators on this project include the following organizations:
- The Institute for Bird Populations (IBP) created and coordinates four landbird monitoring programs that utilize mist netting and banding:
- Monitoring Avian Productivity and Survivorship (MAPS) – breeding season monitoring across North America
- Monitoreo de Sobrevivencia Invernal (MoSI) – winter monitoring in Mexico, Latin America, and the Caribbean
- Monitoring Avian Winter Survival (MAWS) – winter monitoring in the southern U.S.
- Molt Migration Stopover (MoMS) – monitoring of molt-migrant species in the North American Monsoon region
- The Landbird Monitoring Network of the Americas (LaMNA), coordinated by the U.S. forest service redwood science lab.
- UCLA’s High Speed, High Volume Laboratory Network for Infectious Diseases
- Los Alamos National Laboratory
- UCLA School of Public Health, Department of Epidemiology
- Johns Hopkins Bloomberg School of Public Health
- NIH/NIAID Laboratory of Infectious Diseases
- U.S. Department of Agriculture (USDA) Southeast Poultry Research Laboratory
Related Publications